12 May
2017
How to determine the concentration and purity of a DNA sample?
Reliable measurement of DNA concentration and purity is important for many applications in molecular biology where accurate determination of DNA concentration is critical. Impurities in DNA may lead to inaccurate measurement of DNA concentration and could potentially inhibit subsequent labelling reactions.
One of the common methods of measuring nucleic acid purity and concentration are spectrophotometric absorbance of a sample at 260nm compared to the value measured at 280nm . Most scientists also look at the ratios of the 230nm and 260nm absorbance value as an important secondary measure of purity prior using samples in costly and time-constraint downstream applications.
A260/280 Nucleic Acid Ratios
The ratio of absorbance at 260 and 280nm is used to assess the purity of DNA and RNA. A ratio of ~1.8 is generally accepted as “pure” for DNA; a ratio of ~2.0 is generally accepted as “pure” for RNA. If the ratio is appreciably lower in either case, it may indicate the presence of protein, phenol or other contaminants that absorb strongly at or near 280nm.
The following table indicates a general acceptable range of these ratios and does not assure sample purity.
| Sample Type | Ideal | High | Low |
| DNA | ~1.8 | >2.0 | <1.7 |
| RNA | ~2.0 | >2.2 | <1.9 |
| Possible contamination | – | Basic | Acidic Phenol or Protein |
A260/230 Nucleic Acid Ratios
This is a secondary measure of nucleic acid purity. It is used to indicate the presence of unwanted organic contaminants such as Trizol, phenol, Guanidine HCL and guanidine thiocyanate.
Expected 260/230 values are commonly in the range of 1.8-2.2. If the ratio is out of the general acceptable range, it may indicate the presence of contaminants which absorb at 230 nm.
Troubleshooting Purity Ratios
The three most common causes of abnormal 260/280 ratios are described below:
- Samples are very dilute and concentrations are adjacent to the lower detection limit. Abnormal ratios may be observed for nucleic acids samples with concentrations <10ng/µl.
- Sample contamination by residual guanidine, phenol, and other reagents used in the isolation protocol. Carbohydrates are a common contaminant related to nucleic acid isolations from plant sources.
- An inappropriate solution was used for the Blank solution. The Blank solution should be of a similar ionic strength and pH as the sample solution. Eg. Using water for the Blank measurement for samples dissolved in TE may result in low 260/230 ratios.
An example of a nucleic acid sample measured using ND-2800 NanoDOT Nucleic Acid Analyzer are appended in the figures below:
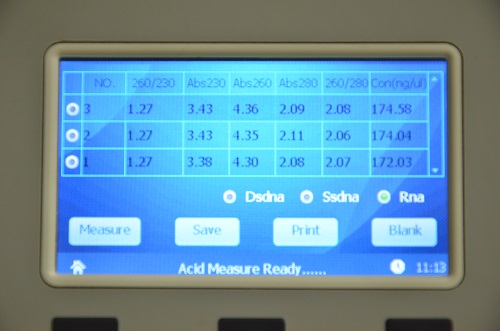
Figure 1. A260/230 ratio, A260/280 ratio and concentration of RNA samples measured using ND-2800 NanoDOT Nucleic Acid Analyzer